tkipf/gcn
{ "createdAt": "2016-11-11T10:59:21Z", "defaultBranch": "master", "description": "Implementation of Graph Convolutional Networks in TensorFlow", "fullName": "tkipf/gcn", "homepage": "", "language": "Python", "name": "gcn", "pushedAt": "2023-04-14T08:09:28Z", "stargazersCount": 7354, "topics": [], "updatedAt": "2026-01-18T11:12:16Z", "url": "https://github.com/tkipf/gcn"}Graph Convolutional Networks
Section titled “Graph Convolutional Networks”This is a TensorFlow implementation of Graph Convolutional Networks for the task of (semi-supervised) classification of nodes in a graph, as described in our paper:
Thomas N. Kipf, Max Welling, Semi-Supervised Classification with Graph Convolutional Networks (ICLR 2017)
For a high-level explanation, have a look at our blog post:
Thomas Kipf, Graph Convolutional Networks (2016)
Installation
Section titled “Installation”python setup.py installRequirements
Section titled “Requirements”- tensorflow (>0.12)
- networkx
Run the demo
Section titled “Run the demo”cd gcnpython train.pyIn order to use your own data, you have to provide
- an N by N adjacency matrix (N is the number of nodes),
- an N by D feature matrix (D is the number of features per node), and
- an N by E binary label matrix (E is the number of classes).
Have a look at the load_data() function in utils.py for an example.
In this example, we load citation network data (Cora, Citeseer or Pubmed). The original datasets can be found here: http://www.cs.umd.edu/~sen/lbc-proj/LBC.html. In our version (see data folder) we use dataset splits provided by https://github.com/kimiyoung/planetoid (Zhilin Yang, William W. Cohen, Ruslan Salakhutdinov, Revisiting Semi-Supervised Learning with Graph Embeddings, ICML 2016).
You can specify a dataset as follows:
python train.py --dataset citeseer(or by editing train.py)
Models
Section titled “Models”You can choose between the following models:
gcn: Graph convolutional network (Thomas N. Kipf, Max Welling, Semi-Supervised Classification with Graph Convolutional Networks, 2016)gcn_cheby: Chebyshev polynomial version of graph convolutional network as described in (Michaël Defferrard, Xavier Bresson, Pierre Vandergheynst, Convolutional Neural Networks on Graphs with Fast Localized Spectral Filtering, NIPS 2016)dense: Basic multi-layer perceptron that supports sparse inputs
Graph classification
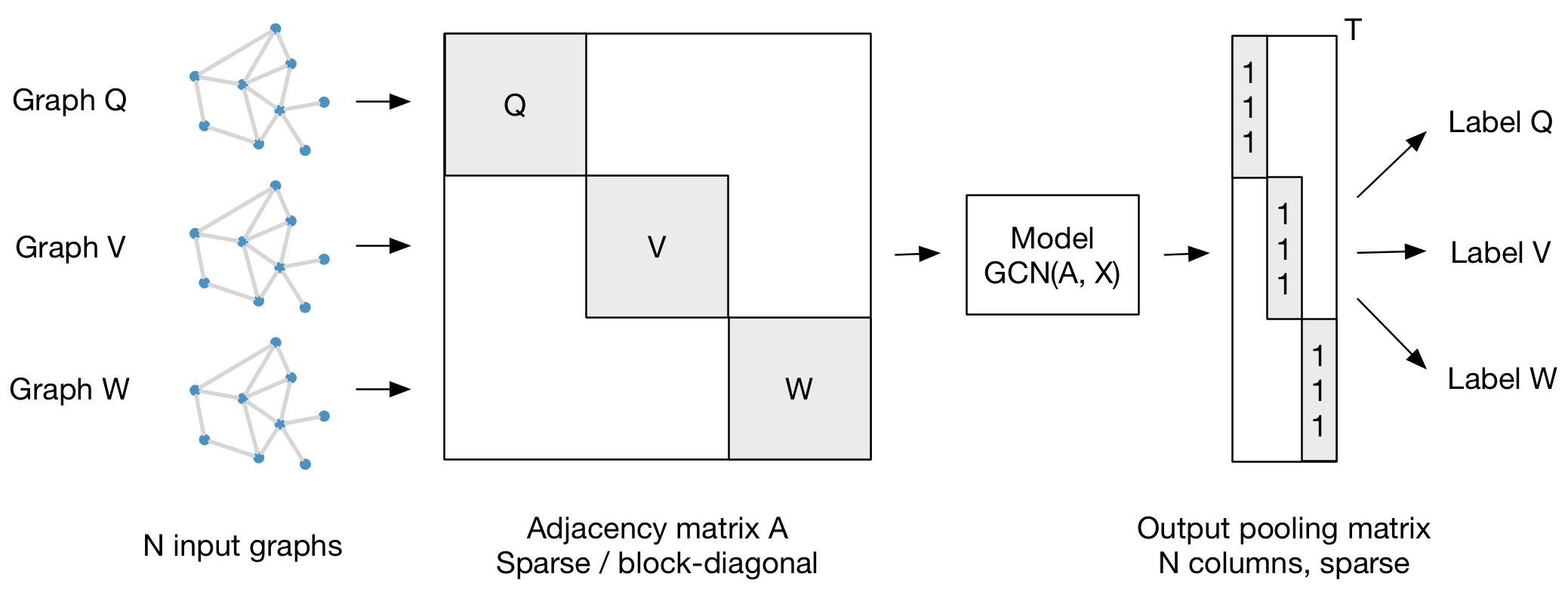
Section titled “Graph classification”Our framework also supports batch-wise classification of multiple graph instances (of potentially different size) with an adjacency matrix each. It is best to concatenate respective feature matrices and build a (sparse) block-diagonal matrix where each block corresponds to the adjacency matrix of one graph instance. For pooling (in case of graph-level outputs as opposed to node-level outputs) it is best to specify a simple pooling matrix that collects features from their respective graph instances, as illustrated below:
Please cite our paper if you use this code in your own work:
@inproceedings{kipf2017semi, title={Semi-Supervised Classification with Graph Convolutional Networks}, author={Kipf, Thomas N. and Welling, Max}, booktitle={International Conference on Learning Representations (ICLR)}, year={2017}}